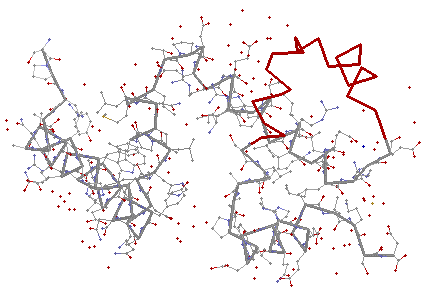
TRP Repressor of E. Coli
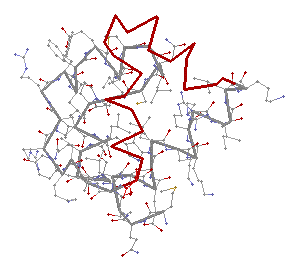
Cro protein of Phage 434
The comparison of the two previous proteins ends up with one principal common substructure which is, as expected, the Helix-Turn-Helix motif, responsible for the binding to the DNA.

|

|
|
TRP Repressor of E. Coli |
Cro protein of Phage 434 |
The registration of the HTH motifs in the two proteins shows that not only is the backbone very well matched, but collateral chains are also pretty well conserved.
The use of frames instead of just points allows the algorithm to discard several false matches of amino acids (4 in this case). Moreover, it makes the algorithm robust with respect to parameters that tune the researched quality of the matches (i.e. the uncertainty that we fix on frames on input).